Novel Method for the Analysis of Single Bio-Molecular Aggregation

Many diseases arise from abnormal molecular aggregation and uncontrolled biomolecular growth or division. Some of the most challenging examples include Alzheimer’s disease, cancer, and angiocardiopathies. Detecting these processes at their earliest stages is critical, as it opens the door to earlier diagnosis and personalised treatments before clinical symptoms appear.
One promising route lies in monitoring microenvironmental changes in biological fluids—such as protein and lipid aggregation or DNA mutations—that often precede disease onset.
The Current Landscape of Pathology Characterization
At present, disease characterisation and assays rely heavily on spectroscopic and microscopy-based techniques. Methods such as:
- Super-resolution and fluorescence microscopy
- Two-photon microscopy
- Raman spectroscopy
- NMR
- FTIR
These approaches often use biomarkers to label molecular indicators, making it possible to visualise and quantify anomalous molecular changes.
While progress has been substantial, these methods still face limitations:
- Difficulty in detecting biomolecular aggregates at ultra-low concentrations
- Inability to provide real-time molecular dynamics in continuously flowing living systems
A New Approach: SHG Integrated with Optical Trapping
At Institut Lumière Matière, Université Claude Bernard Lyon 1, France, I am working within the Brevet group on a system that integrates Second Harmonic Generation (SHG) analysis with optical trapping.
Why SHG?
SHG is a second-order nonlinear optical process that occurs in non-centrosymmetric environments. In this process, two photons at a fundamental frequency (ω) combine, producing a single photon at the harmonic frequency (2ω).
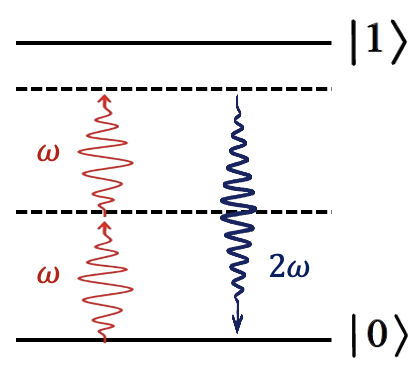
At molecular interfaces, where inversion symmetry is broken, SHG signals are highly sensitive to:
- Molecular symmetry
- Surface defects
- Environmental organisation
This makes SHG a label-free, highly sensitive probe of biomolecular structures—beyond the diffraction limit.
From Ensemble to Individual Molecule Characterisation
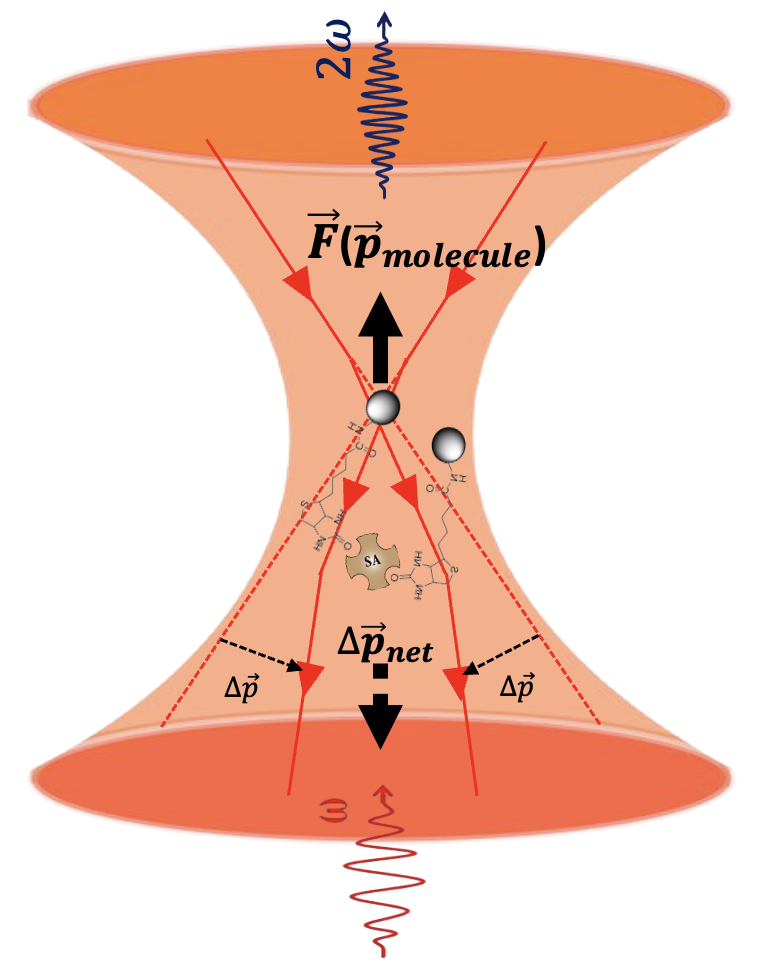
In conventional SHG experiments, measurements reflect only ensemble-averaged values. However, when SHG is performed on a trapped molecule, individual characterisation becomes possible. This breakthrough enables exploration of previously hidden aspects of biomolecular aggregation, such as:
- Biomolecular binding events
- Changes in molecular orientation during particle–particle contact
Pushing the Boundaries Further
To achieve stronger electric field gradients and trap even smaller molecules, in the second year of this PhD study, I will explore an alternative optical trapping method using optical waveguides at RMIT University, Australia.
This complementary approach may unlock new ways of probing molecular interactions at scales and sensitivities not yet accessible.
Outlook
By combining SHG with advanced optical trapping methods, this research aims to provide a powerful, label-free toolkit for studying biomolecular dynamics at the single-molecule level. Such techniques could eventually transform our ability to diagnose, monitor, and understand diseases that originate from abnormal molecular aggregation.
Find out more about my research project here.
Categories
Latest posts

My first year of the PhD as an AI-assisted condition monitoring researcher
Beyond the Science: What I Learned About Research Emissions at the CBH Summer School

From curiosity to research: My path into AI-enhanced cybersecurity
